| Clarify.ai | Helps with out of focus images. Takes a very long time. |
 |  |
| Denoise.ai | Helps to denoise noisy images. Designed for static images without objects moving in between frames. |
 |  |
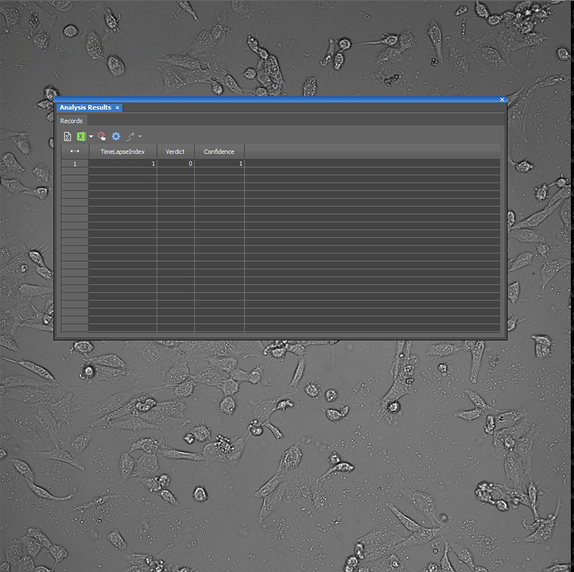
| Cells Presence.ai | Looks for the presence of cells in a brightfield image. Outputs a table with the verdict (0=no cells, 1=cells) and confidence (1=most confident). Was confidently incorrect on the test image.
 |
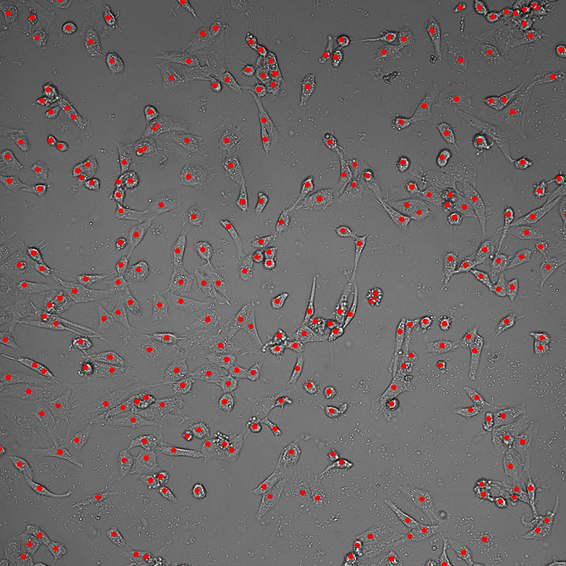
| Cells Localization.ai | Puts a binary layer of dots on cell centers. Must be a brightfield image. 
|
| Enhance.ai | Enhances images with low signal. Neural network must be trained first, then selected. Can train AI using the NIS.ai tab at the top of the NIS Elements window. 
|
| Convert.ai | Once trained, allows for one channel to be converted to another (ex. using FITC data to estimate a DAPI signal). Neural network must be trained first, then selected. Can train AI using the NIS.ai tab at the top of the NIS Elements window. |
| Segment.ai | Using a trained neural network selected from a file, segments objects using a binary layer. Training data requires well-segmented example images. Can train AI using the NIS.ai tab at the top of the NIS Elements window. |
| Segment Objects.ai | Similar to Segment.ai but separates circular objects from each other. Trained neural network must be selected from a file. Can train AI using the NIS.ai tab at the top of the NIS Elements window. |
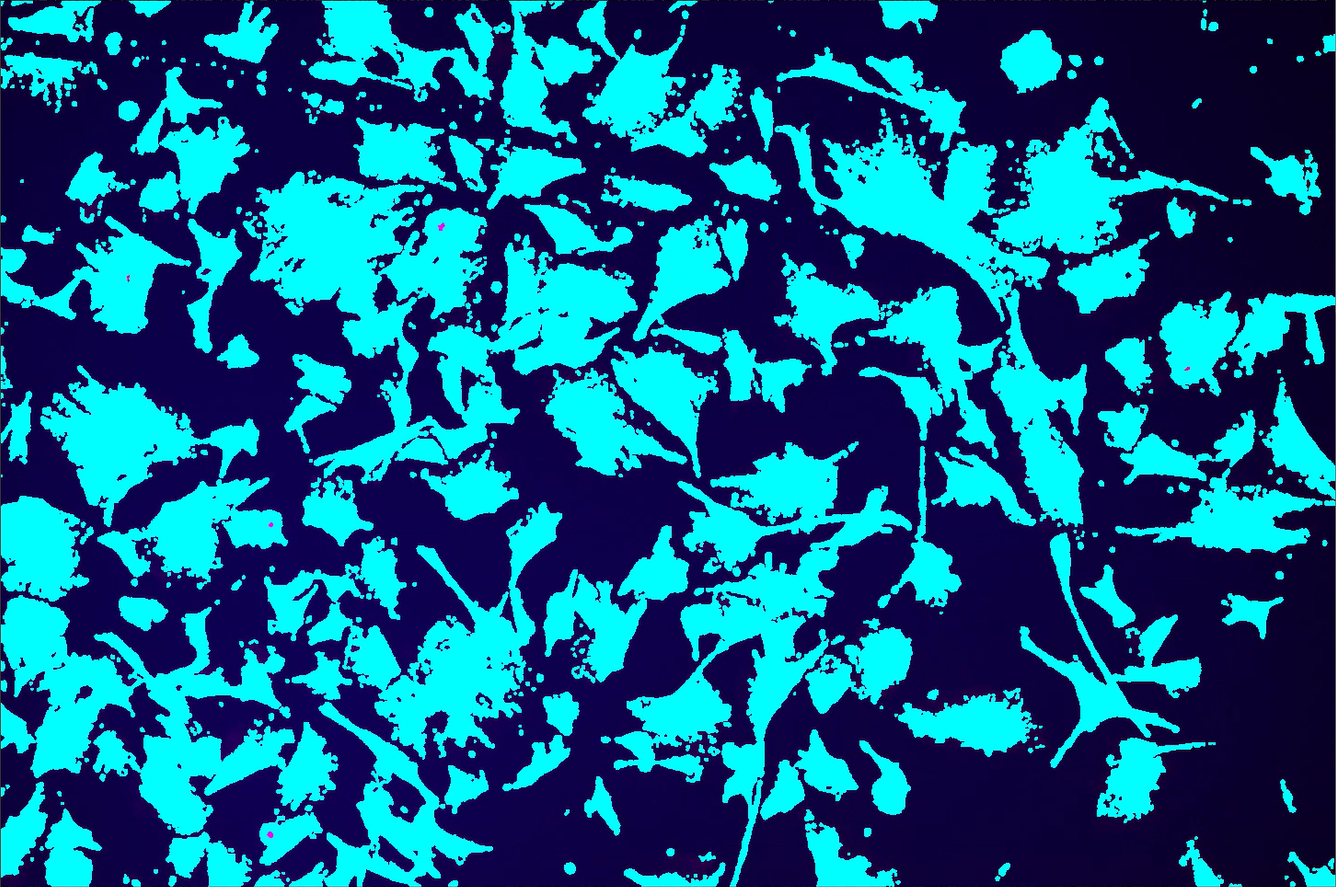
| Homogenous Area/Cells.ai | Looks for homogenous areas and segments image into regions with cells ({0} value) and without cells ({1} value). Does not require selecting a trained file and works pretty fast. Binary can be inverted to switch the cells to {1} values. Example inverted. I (the intern writing this guide) was impressed with how well it worked: like Otsu, but with built-in postprocessing. |
|  |
| Select Trained File.ai | Gives filepaths to trained neural networks. Supposed to be used as an input to other AI nodes, but others do not take table inputs. Also includes various magnifications so an AI file matching the image magnification can be selected. |
| Segmentation Accuracy | Using a binary input for ground truth and another binary input for prediction, creates a table listing number of true positive pixels, false positive pixels, precision (TP/all positives), recall (TP/all pixels that should have been positives), and F1 (2 x precision x recall / (precision + recall)). The IoU is the intersection of union, a quantitative measure of layer overlap. It is found by the overlapping area divided by the union of area: (layer A AND layer B)/ ((layer A OR layer B) – (layer A AND layer B)). |
| Object Segmentation Accuracy | Using a binary input for ground truth and another binary input for prediction, creates a table listing number of true positive objects, false positive objects, precision (TP/all positives), recall (TP/all things that should have been positives), and F1 (2 x precision x recall / (precision + recall)). If the intersection of union between a ground truth object and a prediction object is greater than 0.5, it is considered a true positive. |